Brain Anatomy Segmentation
At Qure.ai, we are getting very good at automatically identifying tumours and lesions in brains from MRI scans. In fact, our team was placed third in Brain Tumour Segmentation (BRATS) challenge at MICCAI 16. As a next step, we wanted to train our machines to understand the anatomy of brain as the prognosis and symptoms of a brain lesion depend upon its anatomical location.
Brain anatomy segmentation will also allow us to do quantitative and morphometric research in neuroimaging. For example, some studies [4, 5] have found that abnormal shapes/volumes of certain anatomical regions are associated with brain disorders like Alzheimer’s disease and Parkinson’s. In fact, there’s a whole subfield called brain morphometry which is concerned with quantifying anatomical features to understand the brain development, aging and diseases.
Brain Anatomy Segmentation is a well-studied problem by now. A MICCAI challenge was held in 2012 to assess the algorithms on whole brain labeling. The challenge provided 15 T1-weighted structural MRI images and associated manually labeled volumes with one label per voxel. These manually marked volumes are called ‘atlases’. 134 anatomical regions were marked in total. Top performer in the challenge [1] achieved a mean dice score of 0.782.
Almost all the brain segmentation algorithms use what is called Multi-atlas Segmentation algorithm. I briefly describe the algorithm [2] below. An illustration follows the description.
Given, Training image-atlas pairs \( (X_i, Y_i), i = 1,2,…,n \) and an unseen test image \( X_{test} \), do:
- Align/Register each of the \(n\) training images \(X_i\) to \(X_{test}\).
- Transform the atlases \(Y_i\) based on the above registrations to the \(X_{test}\) image space to obtain \(Y^i_{pred}, i = 1,2,..,n \).
- Use Majority voting at each voxel or some other similar heuristic (called label/decision fusion) to fuse the \(n\) predictions \(Y^i_{pred}\) and create the final labels \(Y_{pred}\).
Multi-atlas Segmentation Algorithm
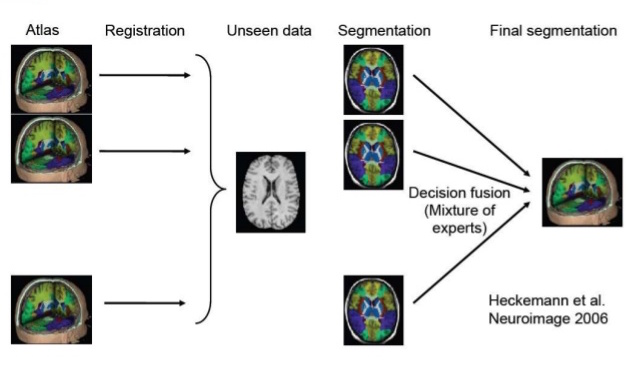
Illustration of Multi-atlas Segmentation (taken from here)
Key steps in Multi-atlas Segmentation are Registration and Label/Decision Fusion. Multiple algorithms are available to perform these steps. Registration is usually non-rigid/deformable. Meaning, small local deformations are made apart from global transformations like rotation and scaling (illustration below).
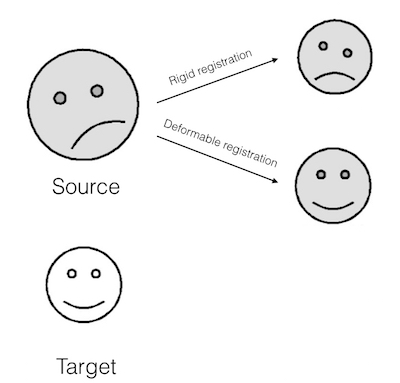
Illustration of Deformable Registration (smileys taken from here)
Rather than selecting the most frequent label at a voxel (Majority voting), an intensity based joint label fusion algorithm [1] can be used to improve the segmentation results. A survey of Multi-atlas Segmentation can be found in [2]. Mature opensource libraries are available for registration and multi atlas segmentation. In the Appendix, I’ll illustrate use of one such library called ANTs and use a subset of MICCAI12 dataset.
A big caveat of Multi-atlas Segmentation is its computational cost. Deformable registration is computationally very expensive. And for a given test image, we’ve to register each training image to it. Label fusion algorithms can also be quite expensive. All of this adds up to 2 or 3 hours for each test image on a latest high-end machine.
This computational cost makes it diffcult to deploy anatomical segmentations for clinical applications. Further research should focus on improving the speed of the segmentation.
One promising research direction is to use deep learning. A deep learning approach to the brain anatomy segmentation [3] was published recently. As noted in [3], main problem is the size of the image and GPU memory. We cannot simply fit a whole MRI image and use 3D CNN to segment because it exceeds the memory available in any single GPU. [3] uses a ‘2.5D’ patch-based segmentation approach; apart from small 3D patch, they also capture the global context by three orthogonal patches, each extracted from the sagittal, coronal and transverse planes.
This model achieves mean dice score of 0.725. This is comparable to best performer [1] in MICCAI12 (0.782), but not as good. This model is also not the easiest to implement. We are working on a fully convolutional 3D CNN approach like U-Net [6] to segment the brain anatomical regions. One of our approaches is to employ model parallelism like in Alex Krizhevsky’s One weird trick paper [7].
References
- Wang, Hongzhi, and Paul Yushkevich. “Multi-atlas segmentation with joint label fusion and corrective learning — an open source implementation.” Frontiers in neuroinformatics 7 (2013): 27
- Iglesias, Juan Eugenio, and Mert R. Sabuncu. “Multi-atlas segmentation of biomedical images: a survey.” Medical image analysis 24.1 (2015): 205-219.
- de Brebisson, Alexander, and Giovanni Montana. “Deep neural networks for anatomical brain segmentation.” Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops. 2015.
- Hutchinson, Michael, and Ulrich Raff. “Structural changes of the substantia nigra in Parkinson’s disease as revealed by MR imaging.” American journal of neuroradiology 21.4 (2000): 697-701.
- Petrella, Jeffrey R., R. Edward Coleman, and P. Murali Doraiswamy. “Neuroimaging and Early Diagnosis of Alzheimer Disease: A Look to the Future 1.” Radiology 226.2 (2003): 315-336.
- Ronneberger, Olaf, Philipp Fischer, and Thomas Brox. “U-net: Convolutional networks for biomedical image segmentation.” International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer International Publishing, 2015.
- Krizhevsky, Alex. “One weird trick for parallelizing convolutional neural networks.” arXiv preprint arXiv:1404.5997 (2014).
Appendix/Code
I’ve created a repo with a script and MRI images. To run this, build ANTs library on your linux/mac machine:
git clone git://github.com/stnava/ANTs.git
mkdir ANTs/bin
cd ANTs/bin
cmake ..
make -j 4 # This might take a while
cp ../Scripts/* bin
# Mac users change bashrc to bash_profile
echo export ANTSPATH=`pwd`/bin/ >> ~/.bashrc
source ~/.bashrc
Once ANTs is installed, run the following commands:
git clone https://github.com/qureai/Multi-Atlas-Segmentation.git
cd Multi-Atlas-Segmentation
bash multi-atlas-segmentation.sh
This should create a file predictions/1004_predLabels.nii.gz which are the predicted anatomical labels for test/1004_3.nii.